|
|
Genetic code
- Central Dogma: DNA -> RNA -> protein
- DNA: resides in the nucleus. It codes information in genes.
- Transcription: Inside the nucleus, the DNA genes get transcribed into RNA (messenger RNAs or mRNAs).
- RNA: The mRNAs get transported out of the nucleus into the cytoplasm. mRNAs are working copies of the gene.
- Translation: ribosomes read off the mRNAs to make proteins.
- Protein: synthesized by ribosomes. They are the end product of what's encoded in the genes and they perform all the functions in the cell.
- The triplet code
- mRNA (nucleotides) must code for protein (amino acids)
- To do this: 3 nucleotide = 1 codon = 1 amino acid
- Codons are continuous, non-overlapping and degenerate
- Continuous because one codon follows right after another. There're no nucleotides in between
- Non-overlapping because the 3 nucleotides that consist of one codon never serve as part of another codon
- Degenerate because more than one codon codes for a given amino acid
- Codon-anticodon relationship
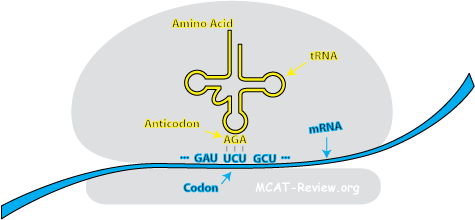
- Anticodon: the 3 bases on the "tip" of the tRNA. A single tRNA contains a single anticodon at the "tip" and the corresponding amino acid at the "tail". Anticodons are complementary to their corresponding codon.
- The codon-anticodon relationship: During translation, codons pair with anticodons so that the correct amino acids can be linked to a given codon.
- Degenerate code, wobble pairing
- Degenerate = more than one codon codes for a given amino acid
- Wobble pairing = the last/3rd nucleotide of the codon doesn't have to match exactly with the anticodon, will still incoporate the same amino acid during translation
- Wobble pairing is the mechanism that explains the degenerate nature of the triplet code
- Missense, nonsense codons
- Missense codon: mutated codon that results in a different amino acid
- Nonsense codon: mutated codon that results in something other than an amino acid. For example, a stop codon
- Initiation, termination codons (function, codon sequences)
- Initiation codon (AUG): signals the start of translation. Lies just downstream of the Shine Dalgarno sequence (Kozak sequence for eukaryotes)
- Termination codon (UAG,UGA,UAA): signals the end of translation. Unlike other codons, tRNA are not involved. Instead a protein called "release factor" comes along and terminates translation
- mRNA composition and structure (RNA nucleotides, 5' cap, poly-A tail)
- mRNA stands for messenger RNA. It's the product of transcription and the template for translation.
- The 5' cap is a modified nucleotide linked in a special way to the mRNA. This protects the 5' end from exonuclease degradation.
- The poly-A tail protects the 3' end of the mRNA from exonuclease degradation.
- Eukaryotic mRNA: 5' cap - nucleotides - 3' polyA.
- Prokaryotic mRNAs don't have the 5' cap or polyA tail.
- More mRNA = more level of expression = more protein made
- Stronger promoter, less DNA/histone methylation = more mRNA made
Transcription
- tRNA, rRNA composition and structure (eg., RNA nucleotides)
- Both tRNA (transfer RNA) and rRNA (ribosomal RNA) are products of transcription. However, they do not serve as the template of translation. tRNA is responsible for bringing in the correct amino acid during translation. rRNA makes up the ribosome, which is the enzyme responsible for translation.
- tRNA is made of nucleotides, many of which is modified for structural and functional reasons. At the 3' end of the tRNA, the amino acid is attached to the 3'OH via an ester linkage.
- tRNA structure: clover leaf structure with anticodon at the tip, and the amino acid at the 3' tail.
- rRNA is made of nucleotides, many of which is modified for structural and functional reasons.
- rRNA is highly structured because it contains the active site for catalysis. The rRNA of the large ribosomal subunit is responsible for catalyzing peptide bond formation, and can do this even without ribosomal proteins.
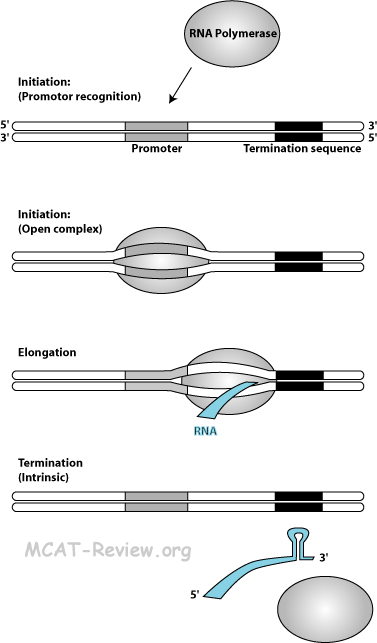
- Mechanism of transcription (RNA polymerase, promoters, primer not required)
- Chain Initiation: RNA polymerase binds to the promoter (TATA box) of the double stranded DNA (closed complex). The double stranded DNA template opens up (open complex).
- Chain elongation: nucleoside triphosphates (AUGCs) adds corresponding to the DNA template. No primer is required. RNA elongates as the RNA polymerase moves down the DNA template. RNA is made from the 5' to 3' direction.
- Chain termination: there are 2 ways that transcription can terminate.
- Intrinsic termination: specific sequences called a termination site creates a stem-loop structure on the RNA that causes the RNA to slip off the template.
- Rho (ρ) dependent termination: a protein called the ρ factor travels along the synthesized RNA and bumps off the polymerase.
- mRNA processing in eukaryotes, introns, exons
- protect from degradation: 5' cap and 3' polyA added
- RNA splicing: cut out introns, keep and stitch together the exons
- Alternate splicing = different ways you can do RNA splicing to make different end products
- Ribozymes, spliceosomes, small nuclear ribonucleoproteins (snRNPs), small nuclear RNAs (snRNAs)
- Ribozymes = RNA enzymes = can have protein parts, but it's actually the RNA part that's doing the catalysis. Best example is the ribosome.
- Spliceosomes = machinery that does RNA splicing = composed of RNA and proteins = possibly a ribozyme
- snRNPs = RNA + protein = subunits that assemble into the spliceosomes and other RNA modification machinery
- snRNAs = the RNA portion of snRNPs
- Functional and evolutionary importance of introns
- Function: RNA splicing, alternate splicing, gene regulation
- Evolution: evolved from selfish/parasitic/mobile genetic elements
Translation
- Roles of mRNA, tRNA, rRNA
- mRNA (messenger RNA): contains codons that code for the peptide sequence.
- tRNA (transfer RNA): contains the anticodon on the "tip" and the corresponding amino acid on the "tail". Link the correct amino acid to its corresponding mRNA codon through codon-anticodon interaction.
- rRNA (ribosomal RNA): forms the ribosome. Catalyzes the formation of the peptide bond.
- Role and structure of ribosomes
- Ribosome is the enzyme that catalyzes protein synthesis.
- Ribosome has 2 subunits - the large and the small.
- The large subunit is responsible for the peptidyl transfer reaction.
- The small subunit is responsible for the recognizing mRNA and binds to the Shine-Dalgarno sequence on the mRNA (Kozak sequence for eukaryotes).
- Both subunits are needed for translation to occur and they come together in a hamburger fashion that sandwiches the mRNA and tRNAs in between.
- Mechanism of translation: initiation, termination, co-factors
- Chain Initiation: To begin translation, you need to form the initiation complex. The initiation complex is basically an assembly of everything needed to begin translation. This includes mRNA, initiator tRNA (fmet), and the ribosome (initiation factors, and GTP aids in the formation of the initiation complex). The initiation complex forms around the initiation codon (AUG), which is just down stream of the Shine-Dalgarno sequence. The Shine-Dalgarno sequence is the "promoter" equivalent of translation for prokaryotes (Kozak sequence for eukaryotes).
- Chain Elongation: protein is made from the N terminus to the C terminus. mRNA codons are read from the 5' to the 3' end. Elongation consists of:
- Binding: new tRNA with its amino acid (tRNA+amino acid is called aminoacyl-tRNA) enters the A site. GTP and elongation factor required.
- Peptidyl transfer: attachment of the new amino acid to the existing chain in the P site. The mechanism is a little strange, what happens is that the already existing chain in the P site migrates and attaches to the aminoacyl-tRNA in the A site.
- Translocation: the lone tRNA in the P site gets kicked off (E site), and the tRNA in the A site, along with the peptide chain attached to it, moves into the P site. The mRNA gets dragged along also - the codon that was in the A site is now in the P site after translocation. The A site is now empty and ready for the binding of a new aminoacyl-tRNA to a new codon. Elongation factor and GTP required.
- Chain termination: When a stop codon is encountered, proteins called release factors, bound to GTP, come in and blocks the A site. The peptide chain gets cleaved from the tRNA in the P site. Peptide chain falls off, and then the whole translation complex falls apart.
- Amino acid activation: enzymes called aminoacyl-tRNA synthetases attach the correct amino acids to their corresponding tRNAs. ATP required.
- Post-translational modification of proteins
- Add carbohydrate groups: localization tags, gives mucous texture to proteoglycans
- Adding fatty groups: membrane anchored proteins
- Acetylation: localization tags
- Disulfide bond formation: joins different subunits
- Phosphorylation: activating/inactivating an enzyme
- Ubiquination: tags protein for destruction
Diagram of translation - graphical overview of initiation, elongation and termination
|
|
